Learning Bayesian Hierarchical Modeling from 8 Schools
The problem we’re discussing in this post appears in Bayesian Data Analysis, 3rd edition (Gelman et al., 2013). Here, Gelman et al. describe the results of independent experiments to determine the effects of special coaching programs on SAT scores.
There are
| A | 28 | 15 |
| B | 8 | 10 |
| C | -3 | 16 |
| D | 7 | 11 |
| E | -1 | 9 |
| F | 1 | 11 |
| G | 18 | 10 |
| H | 12 | 18 |
From BDA3, we consider that the estimates
Non-Hierarchical Methods
Separate estimates
From the table above, we might suspect that schools tend to have different coaching effects – some schools have rather high estimates (like schools A and G), some have small effects (like schools D and F), and some even have negative effects (schools C and E). But the problem is that the standard errors of these estimated effects are very high. If we treat each school as individual experiments and apply separate normal distributions with these values, we see that all of their 95% posterior intervals overlap substantially.
y <- c(28, 8, -3, 7, -1, 1, 18, 12)
sigma <- c(15, 10, 16, 11, 9, 11, 10, 18)
q_025 <- rep(0, 8)
q_975 <- rep(0, 8)
for (i in 1:8){
q_025[i] <- qnorm(0.025, mean = y[i], sd = sigma[i])
q_975[i] <- qnorm(0.975, mean = y[i], sd = sigma[i])
}
print(cbind(y, sigma, q_025, q_975))
y sigma q_025 q_975
[1,] 28 15 -1.39946 57.39946
[2,] 8 10 -11.59964 27.59964
[3,] -3 16 -34.35942 28.35942
[4,] 7 11 -14.55960 28.55960
[5,] -1 9 -18.63968 16.63968
[6,] 1 11 -20.55960 22.55960
[7,] 18 10 -1.59964 37.59964
[8,] 12 18 -23.27935 47.27935
Pooled estimates
The above overlap based on independent analyses seems to suggests that all experiments might be estimating the same quantity. We can take another approach, and that is to treat the given data as eight random sample under a common normal distribution with known variances. With a noninformative prior, it can be shown that the posterior mean and variance is the inverse weighted average of
cat(paste('Posterior mean:', sum(y/sigma^2)/sum(1/sigma^2)), '\n')
cat(paste('Posterior variance:'), 1/sum(1/sigma^2))
Posterior mean: 7.68561672495604
Posterior variance: 16.58053
The
“The pooled model implies the following statement: ‘the probability is 0.5 that the true effect in A is less than 7.7,’ which, despite the non-significant
Ideally, we want to combine information from all of these eight experiments without assuming the
Bayesian Hierarchical Modeling
We can model this dataset as such: the coaching effect
The vector of parameters
With this setup, we can try to combine the coaching estimates in some way to obtain improved estimates of the true effects
We can write an expression for the unnormalized full posterior density
Next, we can decompose the full posterior density into the conditional posterior,
which will be useful in matching the variance part of the normal densities in this decomposition.
where
and
By forming a quadratic expression in terms of
The posterior mean,
Parameter Estimation
The solution is not yet complete, because
Consider a transformed set of parameters
We can try to get a good estimate of
Now we can write the log posterior as
where the last term comes from the Jacobian.
Let’s visualize the log posterior with a contour plot.
# given data
y <- c(28, 8, -3, 7, -1, 1, 18, 12)
sigma <- c(15, 10, 16, 11, 9, 11, 10, 18)
# defining the log posterior for lambda
logpost <- function(lambda, sigma, y){
sum(-0.5*log(exp(2*lambda[2])+sigma^2) -
((lambda[1]-y)^2)/(2*(sigma^2+exp(2*lambda[2])))) +
lambda[2]
}
# grids
lambda_1 <- seq(from = -18, to = 37, by = 0.1)
lambda_2 <- seq(from = -6, to = 4.1, by = 0.1)
z <- matrix(0, nrow = length(lambda_1), ncol = length(lambda_2))
for (i in 1:length(lambda_1)){
for (j in 1:length(lambda_2)){
lambda <- c(lambda_1[i], lambda_2[j])
z[i,j] <- logpost(lambda, sigma, y)
}
}
contour(x = lambda_1, y = lambda_2, z = z, col = "blue", nlevels = 40,
xlab = expression(lambda[1]), ylab = expression(lambda[2]),
cex.axis = 1.1, cex.lab = 1.3)
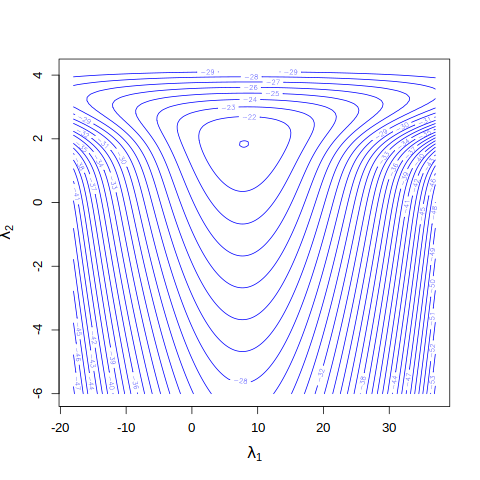
From the contour plot, the mode seems close to optim() to find the posterior mode and covariance matrix by approximating the log posterior to a (multivariate) normal distribution.
out <- optim(par = c(8, 2), fn = logpost, control = list(fnscale = -1),
hessian = TRUE, sigma = sigma, y = y)
cat('Posterior mode:\n')
print((post_mode <- out$par))
cat('\n')
cat('Covariance matrix: \n')
print((post_cov <- -solve(out$hessian)))
Posterior mode:
[1] 7.926685 1.841525
Covariance matrix:
[,1] [,2]
[1,] 22.3232882 0.1935228
[2,] 0.1935228 0.5352576
The normal approximation to the posterior of
The covariance matrix will be useful when sampling for values of
MCMC Sampling
The Metropolis-Hastings (MH) algorithm is a MCMC method to generate random samples from a density where direct sampling might be difficult (e.g. where normalizing constants are intractable or for high dimensional densities). As this post gets rather lengthy, I shall skip the introduction to the MH algorithm or reserve it for future posts.
Here, we will use MH algorithm to draw 10000 samples. We will use our normal approximation density has the proposal here, as it is the closest to our target posterior density and hence it is more likely to generate accepted samples. The first 5000 samples will be treated as burn-in and discarded; desired samples are obtained after the stationary distribution is reached.
library(LearnBayes)
library(coda)
set.seed(11)
iters <- 10^4
proposal <- list(var = post_cov, scale = 2)
# random walk metropolis
fit1 <- rwmetrop(logpost, proposal, start = post_mode, iters, sigma, y)
# overlaying last 5000 draws on contour plot of the log posterior
contour(x = lambda_1, y = lambda_2, z = z, col = "blue", nlevels = 40,
xlab = expression(lambda[1]), ylab = expression(lambda[2]),
cex.axis = 1.1, cex.lab = 1.3)
points(x = fit1$par[5001:iters,1], y = fit1$par[5001:iters,2], col = "red")
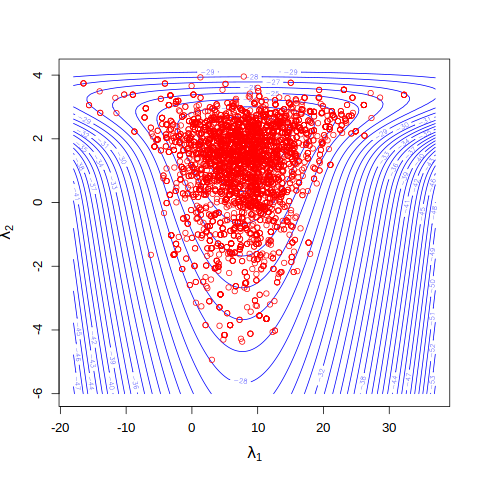
cat('Acceptance rate: \n')
print(fit1$accept)
Acceptance rate:
[1] 0.3288
par(mfrow=c(2,1))
plot(density(fit1$par[5001:iters,1]), main = "", xlab = expression(lambda[1]))
plot(density(fit1$par[5001:iters,2]), main = "", xlab = expression(lambda[2]))
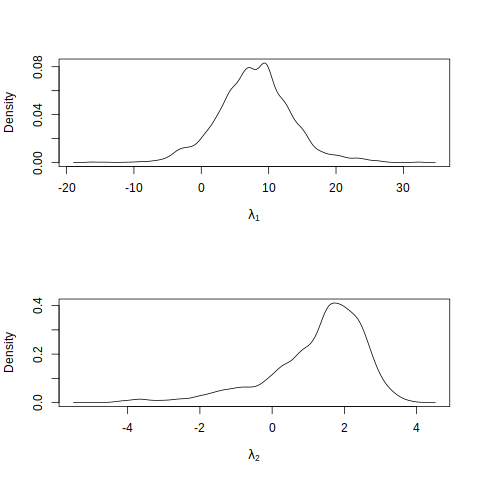
The sampling acceptance rate is 32.88%, which is reasonable, and we observe that the MCMC samples
mcmcobj1 <- mcmc(fit1$par[5001:iters,])
colnames(mcmcobj1) <- c("lambda_1", "lambda_2")
par(mfrow=c(2,1))
traceplot(mcmcobj1)
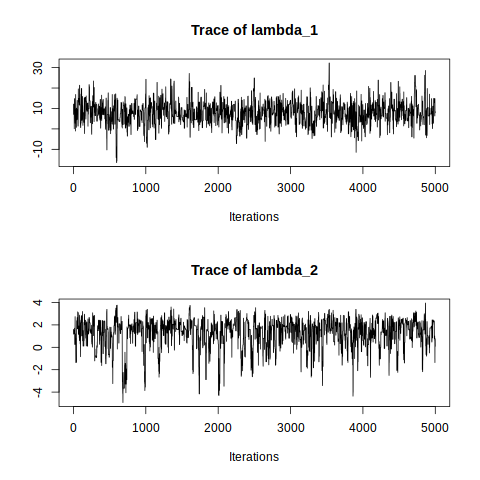
The traceplots of both
It is also important to analyze the degree of autocorrelation in the sampled values. In an MCMC algorithm like the random-walk Metropolis-Hastings above, the simulated value of
par(mfrow=c(2,1))
autocorr.plot(mcmcobj1, auto.layout = FALSE)
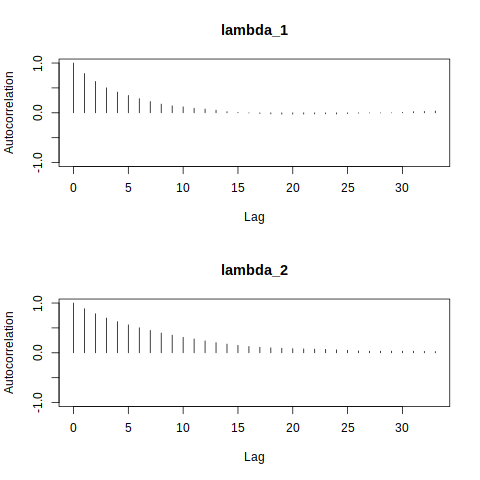
Here, the autocorrelation plots show fast decay in both
With a satisfactory MCMC output analysis, we can use these samples to obtain samples of true effects,
# the last 5000 MCMC samples (lambda_1, lambda_2)
lambda_samples <- fit1$par[5001:iters,]
# function to compute mean
theta_hat <- function(lambda, y_j, sigma_j){
((y_j/sigma_j^2)+(lambda[,1]/exp(2*lambda[,2]))) /
((1/sigma_j^2)+(1/exp(2*lambda[,2])))
}
# function to compute variance
V <- function(lambda, y_j, sigma_j){
1 / (1/sigma_j^2 + 1/exp(2*lambda[,2]))
}
# drawing 5000 samples of theta_j
theta_samples <- function(lambda, y_j, sigma_j){
rnorm(5000, mean = theta_hat(lambda, y_j, sigma_j),
sd = sqrt(V(lambda, y_j, sigma_j)))
}
theta_mean <- rep(0, 8)
theta_sd <- rep(0,8)
# the joint posterior density of (theta_1,...,theta_j)
theta_all <- matrix(0, nrow = 5000, 8)
for (j in 1:8){
thetas <- theta_samples(lambda_samples, y[j], sigma[j])
theta_all[,j] <- thetas
theta_mean[j] <- mean(thetas)
theta_sd[j] <- sd(thetas)
}
print(theta_dist <- cbind(theta_mean, theta_sd))
theta_mean theta_sd
[1,] 11.226786 8.510583
[2,] 7.812253 6.185383
[3,] 6.078697 7.993831
[4,] 7.609353 6.515474
[5,] 5.162853 6.381664
[6,] 6.231208 6.729192
[7,] 10.340858 6.990141
[8,] 8.490497 8.045273
We arrive at estimates of the true coaching effect
Shrinkage
From the conditional posteriors above, we can find that the posterior mean of
where
is the size of the shrinkage of
# shrinkage function for each j
shrink_j <- function(lambda, sigma_j){
(1/exp(lambda[,2]))^2 / ((1/exp(lambda[,2]))^2+1/sigma_j^2)
}
shrink <-rep(0, 8)
for(j in 1:8){
shrink[j] <- mean(shrink_j(lambda_samples, sigma[j]))
}
print(data.frame(school = LETTERS[c(1:8)],
shrink_size = shrink,
rank_shrink =rank(shrink),
rank_sigma = rank(sigma)))
school shrink_size rank_shrink rank_sigma
1 A 0.8328975 6.0 6.0
2 B 0.7376910 2.5 2.5
3 C 0.8458181 7.0 7.0
4 D 0.7620532 4.5 4.5
5 E 0.7096051 1.0 1.0
6 F 0.7620532 4.5 4.5
7 G 0.7376910 2.5 2.5
8 H 0.8676774 8.0 8.0
We observe that shrinkage and sigma values for each school have the same rank. This is consistent with the shrinkage formula above; since the squared inverse of
The samples also provide a way draw other related inferences, such as the probability of seeing an effect as large as 28 for school A, which works out to be a very low value.
sum(theta_all[,1] > 28) / length(theta_all[,1])
0.0468
Note the contrast with the “separate estimates” approach we discussed earlier, which would imply that this probability is 50%, which seems overly large especially given the data from other schools.
We can also ask for the probability that school A has a greater coaching effect than the rest of the schools.
prob <-c()
for(j in 2:8){
prob[j] <-mean(sum(theta_all[,1] > theta_all[,j])) / nrow(theta_all)
}
print(data.frame(school = LETTERS[c(1:8)], probability = prob))
school probability
1 A NA
2 B 0.6346
3 C 0.6800
4 D 0.6382
5 E 0.7162
6 F 0.6804
7 G 0.5382
8 H 0.5994
The probability that school A’s coaching effect is greater than the other schools doesn’t seem that large, even though the original estimates
Conclusion
In summary, Bayesian hierarchical modeling gives us a way to calculate “true effect” sizes that is otherwise hard to obtain (we only have unbiased estimates and standard errors from our dataset). Arguably, the assumptions of both the “separate estimates” and “pooled estimates” approach don’t fully capture the state of our knowledge to be able to use them convincingly. But with the hierarchical model, we now have a “middle ground” of sorts, and it is also flexible enough to incorporate both empirical data and any prior beliefs we might have, both summarized by the posterior distribution. Finally, we can obtain samples using MCMC methods, from which we can perform inferences.
Credits
I learnt of this interesting problem as a piece of assignment from my Bayesian Statistics class, ST4234 in NUS, taught by Prof Li Cheng. I also referred to Bayesian Data Analysis, 3rd edition by Gelman et al for further context and some relevant statistical arguments.
Cover image: Jason Leung (Unsplash)
References
Enjoyed reading this article? More articles you might like to read next: