Understanding Copulas
In statistics, copulas are functions that allow us to define a multivariate distribution by specifying their univariate marginals and interdependencies separately. In modelling returns of assets, for example, this enables greater flexibility and ability to model joint behaviour in extreme events.
Let’s study this in further detail using daily log returns of two assets, Apple and Goldman Sachs, over a 12-year period.
library(tseries)
options("getSymbols.warning4.0"=FALSE)
a <- get.hist.quote(instrument = 'AAPL',
start="2009-01-04", end="2021-01-04",
quote = c("AdjClose"), provider = "yahoo",
compress = "d")
b <- get.hist.quote(instrument = 'GS',
start="2009-01-04", end="2021-01-04",
quote = c("AdjClose"), provider = "yahoo",
compress = "d")
df <- data.frame(list(diff(log(a)), diff(log(b))))
colnames(df) <- c('aapl', 'gs')
time series starts 2009-01-05
time series ends 2020-12-31
time series starts 2009-01-05
time series ends 2020-12-31
Let’s take a peek at the top 10 rows of the dataframe.
print(head(df[1:10,], 10))
aapl gs
2009-01-06 -0.01663156 -0.0007888361
2009-01-07 -0.02184523 -0.0486211860
2009-01-08 0.01839934 0.0107118530
2009-01-09 -0.02313506 -0.0175993365
2009-01-12 -0.02142469 -0.0773947112
2009-01-13 -0.01077318 0.0032132961
2009-01-14 -0.02750955 -0.0290366487
2009-01-15 -0.02311758 -0.0248805384
2009-01-16 -0.01267316 -0.0106212447
2009-01-20 -0.05146606 -0.2102222980
Modeling Tail Dependence
Say we want to estimate tail dependence of these assets, i.e. co-movements at the extreme ends of daily returns. In other words, what is the chance that AAPL’s worst cases are also GS’s worst cases?
Let
We first compare the tail depedencies, at various probabilities, of the empirical data and 100000 samples from a bivariate normal distribution (with its mean and covariance matrix estimated from the data).
# parameter estimates
cat('Sample mean:\n')
cat(df_means <- c(mean(df[,1]), mean(df[,2])))
cat('\n\n')
cat('Sample covariance:\n')
print((df_cov <- cov(df)))
library(mvtnorm)
set.seed(42)
# 100k samples from bivariate normal
mvn_samples <- rmvnorm(1e5, df_means, df_cov)
Sample mean:
0.001264829 0.0004185186
Sample covariance:
aapl gs
aapl 0.0003283744 0.0001778626
gs 0.0001778626 0.0004364080
probs <- c(0.2, 0.1, 0.05, 0.02, 0.01, 0.005, 0.001)
tally1 <- matrix(0, 2, 7)
for (i in 1:7){
q = probs[i]
tally1[,i] = c(
(sum((df[,1]<quantile(df[,1], q))*(df[,2]<quantile(df[,2], q))) /
sum((df[,1]<quantile(df[,1], q)))),
(sum((mvn_samples[,1]<quantile(mvn_samples[,1], q)) *
(mvn_samples[,2]<quantile(mvn_samples[,2], q)))
/ sum((mvn_samples[,1]<quantile(mvn_samples[,1], q))))
)
}
tally1_df <- as.data.frame(tally1, row.names=c('observed','normal'))
colnames(tally1_df) <- as.character(probs)
print(tally1_df)
0.2 0.1 0.05 0.02 0.01 0.005 0.001
observed 0.4668874 0.4337748 0.397351 0.3114754 0.3225806 0.1875 0.50
normal 0.4176500 0.3066000 0.218000 0.1570000 0.1130000 0.0800 0.07
We observe as the probabilities get smaller, the calculated tail dependences between empirical returns and data sampled from the bivariate normal distribution begins to differ greatly.
Let’s try to do better with copulas.
Introducing Copulas
The term ‘copula’ is derived from the Latin for ‘link’, and in our context, is named aptly so. We can understand copulas as multivariate cumulative distribution functions that link marginal distributions and describe their interdependencies. Its marginal distributions are all Uniform(0,1), we use Uniform as a ‘bridge’ since a random variable from any distribution can be transformed to Uniform and back with the probability integral transform.
The copula of a random vector
Some common examples include
- independence copula:
- co-monotonicity copula:
- Gaussian copula:
We will be using the copula package, which has various common predefined copulas for us to choose and sample from.
Estimating Marginal Distributions
Before that, let us first fit marginal distributions for the daily returns of AAPL and GS with the help of the MASS package. fitdistr() will help us find the optimal parameters given a distribution, so let us compare between the AIC for Normal, t and Cauchy distributions.
options(warn=-1)
library(MASS)
cat('AAPL\n')
cat(paste('Normal:\t',AIC(fitdistr(df$aapl, 'normal')),'\n'))
cat(paste('t:\t',AIC(fitdistr(df$aapl, 't')), '\n'))
cat(paste('Cauchy:\t',AIC(fitdistr(df$aapl, 'cauchy')), '\n'))
cat('\nGS\n')
cat(paste('Normal:\t',AIC(fitdistr(df$gs, 'normal')),'\n'))
cat(paste('t:\t',AIC(fitdistr(df$gs, 't')), '\n'))
cat(paste('Cauchy:\t',AIC(fitdistr(df$gs, 'cauchy')), '\n'))
AAPL
Normal: -15645.9234978236
t: -16179.9680136581
Cauchy: -15654.6146727265
GS
Normal: -14787.2501190948
t: -15752.4739791247
Cauchy: -15282.9761802124
t distribution gives the lowest AIC, so we shall use that as our marginals. Let’s proceed to extract the optimal parameters for both assets. Note that fitdistr() uses the location-scale family, so besides the degree of freedom, location m and scale s are returned as well.
cat('AAPL\n')
(aapl_t_param <- fitdistr(df$aapl, 't'))
aapl_m <- aapl_t_param$estimate['m']
aapl_s <- aapl_t_param$estimate['s']
aapl_df <- aapl_t_param$estimate['df']
cat('\nGS\n')
(gs_t_param <- fitdistr(df$gs, 't'))
gs_m <- gs_t_param$estimate['m']
gs_s <- gs_t_param$estimate['s']
gs_df <- gs_t_param$estimate['df']
AAPL
m s df
0.0014079306 0.0122031312 3.4246717881
(0.0002668132) (0.0002885762) (0.2373954742)
GS
m s df
0.0005630129 0.0124256289 2.9726045290
(0.0002772650) (0.0002930408) (0.1810629562)
We’ll now transform the data into Uniform(0,1) by taking their order statistics and dividing it by the number of observations plus one. The ‘+1’ is added as a pseudo-observation so that all variates are forced inside the unit space to avoid problems with density evaluations at the boundaries. Without this, fitcopula() will throw an error.
As a side note, let’s briefly see how this works. We want to show that taking the ranks of variates
With
Then
u_aapl <- rank(df$aapl)/(nrow(df)+1)
u_gs <- rank(df$gs)/(nrow(df)+1)
u_df <- data.frame(list(u_aapl, u_gs))
colnames(u_df) <- c('u_aapl', 'u_gs')
# original density of returns
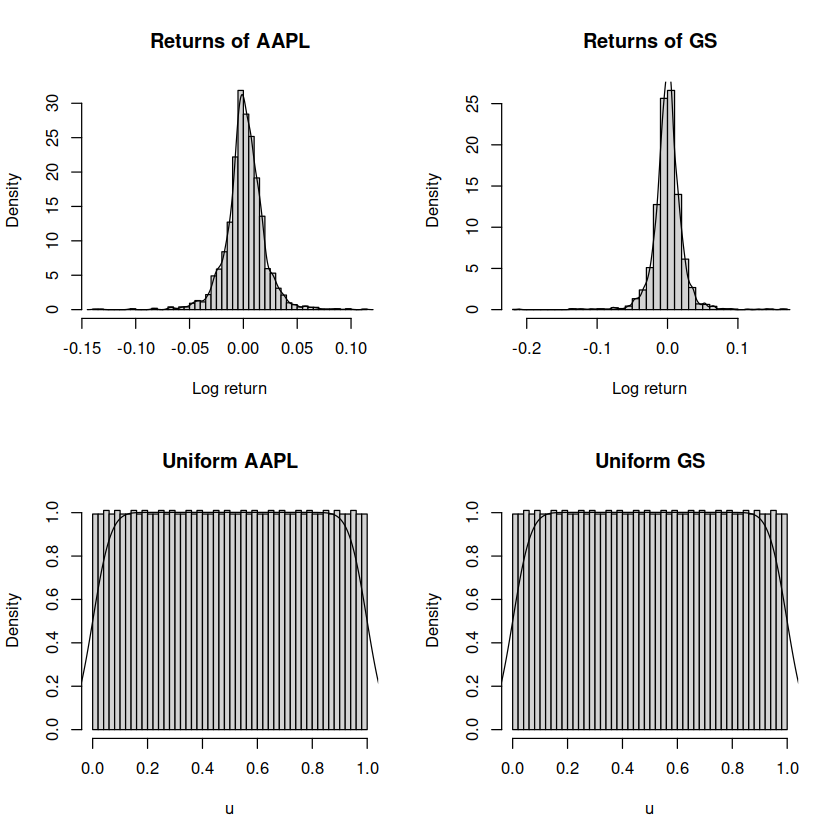
par(mfrow=c(2, 2))
hist(df$aapl, freq=FALSE, breaks=50,
main="Returns of AAPL", xlab="Log return")
lines(density(df$aapl))
hist(df$gs, freq=FALSE, breaks=50,
main="Returns of GS", xlab="Log return")
lines(density(df$gs))
# transformed density of returns (uniform)
hist(u_aapl, freq=FALSE, breaks=50,
main="Uniform AAPL", xlab="u")
lines(density(u_aapl))
hist(u_gs, freq=FALSE, breaks=50,
main="Uniform GS", xlab="u")
lines(density(u_gs))
Choosing and Fitting Copulas
The copula library gives a wide selection of common copulas (elliptical and frequently-used Archimedean copulas). Fitting a few, we observe that the t copula gives us the best fit in terms of maximum pseudo-likelihood.
library(copula)
fitCopula(normalCopula(dim=2), data=u_df)
cat('\n\n')
fitCopula(tCopula(dim=2), data=u_df)
cat('\n\n')
fitCopula(gumbelCopula(dim=2), data=u_df)
Call: fitCopula(copula, data = data)
Fit based on "maximum pseudo-likelihood" and 3019 2-dimensional observations.
Copula: normalCopula
rho.1
0.439
The maximized loglikelihood is 320.9
Convergence problems: code is 52 see ?optim.
Call: fitCopula(copula, data = data)
Fit based on "maximum pseudo-likelihood" and 3019 2-dimensional observations.
Copula: tCopula
rho.1 df
0.4327 4.6111
The maximized loglikelihood is 372
Optimization converged
Call: fitCopula(copula, data = data)
Fit based on "maximum pseudo-likelihood" and 3019 2-dimensional observations.
Copula: gumbelCopula
alpha
1.361
The maximized loglikelihood is 291.7
Optimization converged
A 2-dimensional t-copula has the following form:
where
Let’s fit a t copula with the fitted parameters from above (
Then, again with the probability integeral transform, we transform the these Uniform samples back to their marginal distributions, which we have selected as t distributions as studied earlier. Since the quantile
t_cop_fit_est <- fitCopula(tCopula(dim=2), data=u_df)@estimate
t_cop_fit_rho <- t_cop_fit_est[1]
t_cop_fit_df <- t_cop_fit_est[2]
t_cop <- tCopula(t_cop_fit_rho, df=t_cop_fit_df)
t_cop_samples <- rCopula(1e5, copula=t_cop)
t_cop_aapl <- qt(t_cop_samples[,1], df=aapl_df) * aapl_s + aapl_m
t_cop_gs <- qt(t_cop_samples[,2], df=gs_df) * gs_s + gs_m
Tail Dependence with Copula
With the generated marginal samples, we can now calculate tail depedence using the method we saw earlier.
tally2 <- matrix(0, 3, 7)
for (i in 1:7){
q = probs[i]
tally2[,i] = c(
(sum((df[,1]<quantile(df[,1], q))*(df[,2]<quantile(df[,2], q))) /
sum((df[,1]<quantile(df[,1], q)))),
(sum((mvn_samples[,1]<quantile(mvn_samples[,1], q)) *
(mvn_samples[,2]<quantile(mvn_samples[,2], q)))
/ sum((mvn_samples[,1]<quantile(mvn_samples[,1], q)))),
(sum((t_cop_aapl<quantile(t_cop_aapl, q))*(t_cop_gs<quantile(t_cop_gs, q))) /
sum((t_cop_aapl<quantile(t_cop_aapl, q))))
)
}
tally2_df <- as.data.frame(tally2, row.names=c('observed',
'normal',
't copula'))
colnames(tally2_df) <- as.character(probs)
print(tally2_df)
0.2 0.1 0.05 0.02 0.01 0.005 0.001
observed 0.4668874 0.4337748 0.397351 0.3114754 0.3225806 0.1875 0.50
normal 0.4176500 0.3066000 0.218000 0.1570000 0.1130000 0.0800 0.07
t copula 0.4217500 0.3365000 0.293800 0.2590000 0.2380000 0.2440 0.23
It is also possible to calculate the tail dependence of copulas by
2-2*(pt(sqrt(t_cop_fit_df+1)*sqrt(1-t_cop_fit_rho)/
sqrt(1+t_cop_fit_rho),
df=t_cop_fit_df+1))
0.190010784498546
Although empirically at
Compared to simulated data from the bivariate normal distribution earlier, the simulation from the t copula is closer to the empirical data and produce substantial estimates at the tail, albeit still lower. In extreme cases like
In the event of insufficiency of data, copulas are also able to provide a theoretical measure of tail dependence. It is however noteworthy that not all copulas model tail dependences. t copula provides the above formula for both lower and upper tail dependences, while Gumbel copula, for example, only models upper tail dependence.
Cover image: Karine Avetisyan (Unsplash)
Enjoyed reading this article? More articles you might like to read next: